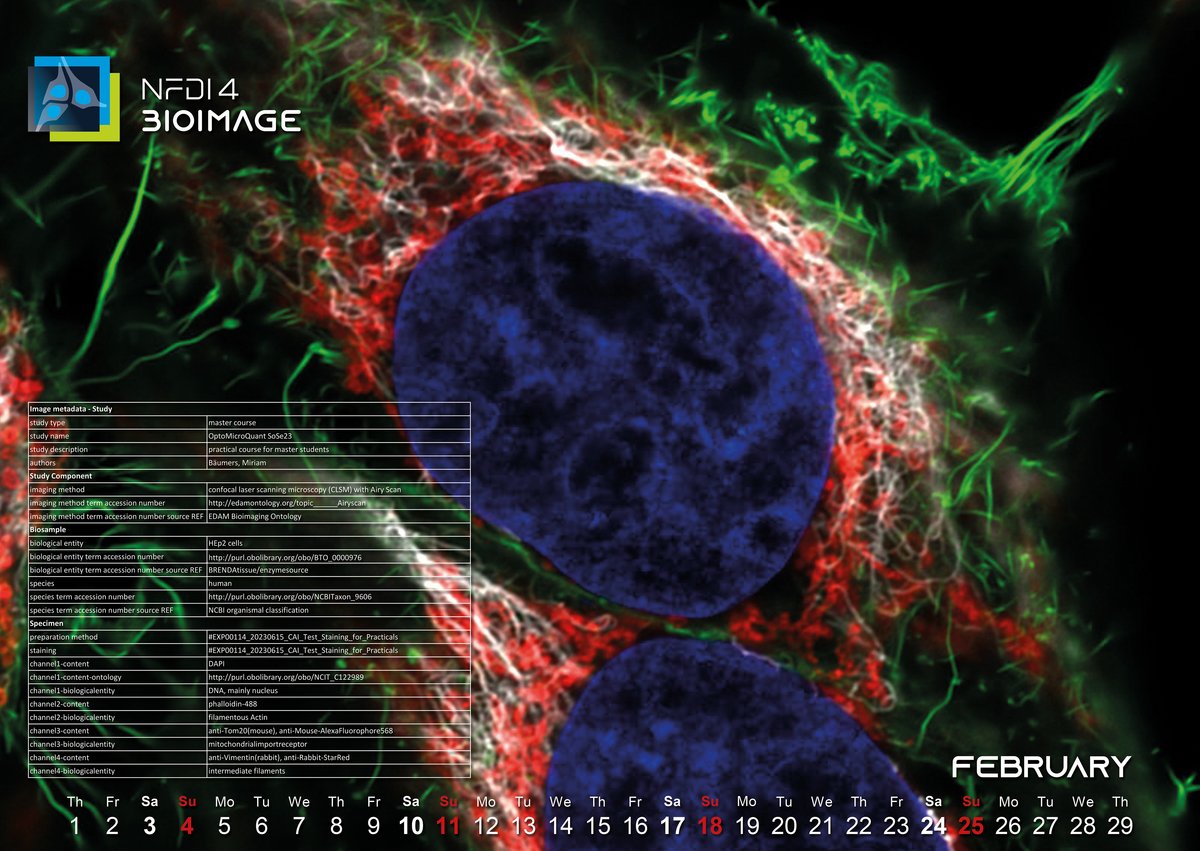
February 2024: The internal wiring of a cancer cell line
The February image was acquired in 2023 during a microscopy course for master students at the Center for Advanced imaging (CAi) at the Heinrich-Heine-University in Düsseldorf. It depicts two HEp2 cells, a cancer cell line used in many laboratories, which after fixation were stained with dyes and antibodies. In blue, you see the nucleus stained with DAPI, green represents the actin cytoskeleton, red depicts the mitochondria and intermediate filaments can be seen in white.
The image is beautiful, but we ask: Is it also FAIR1? The raw file is safely archived on a file server at CAi, but users who would like to examine or re-use the file would need to find the right location, successfully download and interpret the format in which it was written. To improve this situation, the file has also been imported into OMERO (Open Microscopy Environment Remote Object), a client-server software platform for visualizing, managing, and annotating scientific image data2. The data is still stored on a server within CAi but can be accessed from everywhere the user has internet access. In conclusion, the data is Findable and Accessible.
The data is also Interoperable: OMERO uses the Bio-Formats3 tool, a tool that is able to read many proprietary file formats from microscopy vendors, to read microscopic images during import. So even though, the original file is in a proprietary file format, it can either be read by another researcher using the Bio-Formats tool or it can be exported from OMERO using another, more interoperable file format.
The reusability of the image is defined by the amount of (biological) metadata that is attached to the image. The hardware metadata (which microscope was used, which objective, which filters etc.) is read by Bio-Formats during the import and is displayed in the OMERO interface. The biological metadata has to be added manually by the user, which can be done using the annotation tools in OMERO. Here we decided to add all relevant information using key-value pairs.
But how does the user decide which information is relevant and how to structure it? We decided to follow the REMBI (Recommended Metadata for Biological Images) guidelines published by Sarkens et al. in 2021. REMBI suggests categories for biological information (e.g. Study, Biosample, Specimen…) and includes the use of ontologies.
While metadata annotation seems laborious at first, it is an essential step to make biological images reusable. We think that taking the time to make sure your images are reusable and therefore FAIR is time invested in the future: of your work (when preparing for publication eventually), of the image itself and also of the scientific community that might reuse your image to answer yet another scientific question.
Vanessa, Data Steward in the NFDI4BIOIMAGE consortium and at CAi
1: FAIR = Findable, Accessible, Interoperable, Reusable
2: https://omero.readthedocs.io/en/stable/users/index.html
3: https://www.openmicroscopy.org/bio-formats/
4: Sarkans, U., Chiu, W., Collinson, L. et al. REMBI: Recommended Metadata for Biological Images—enabling reuse of microscopy data in biology. Nat Methods 18, 1418–1422 (2021). https://doi.org/10.1038/s41592-021-01166-8
