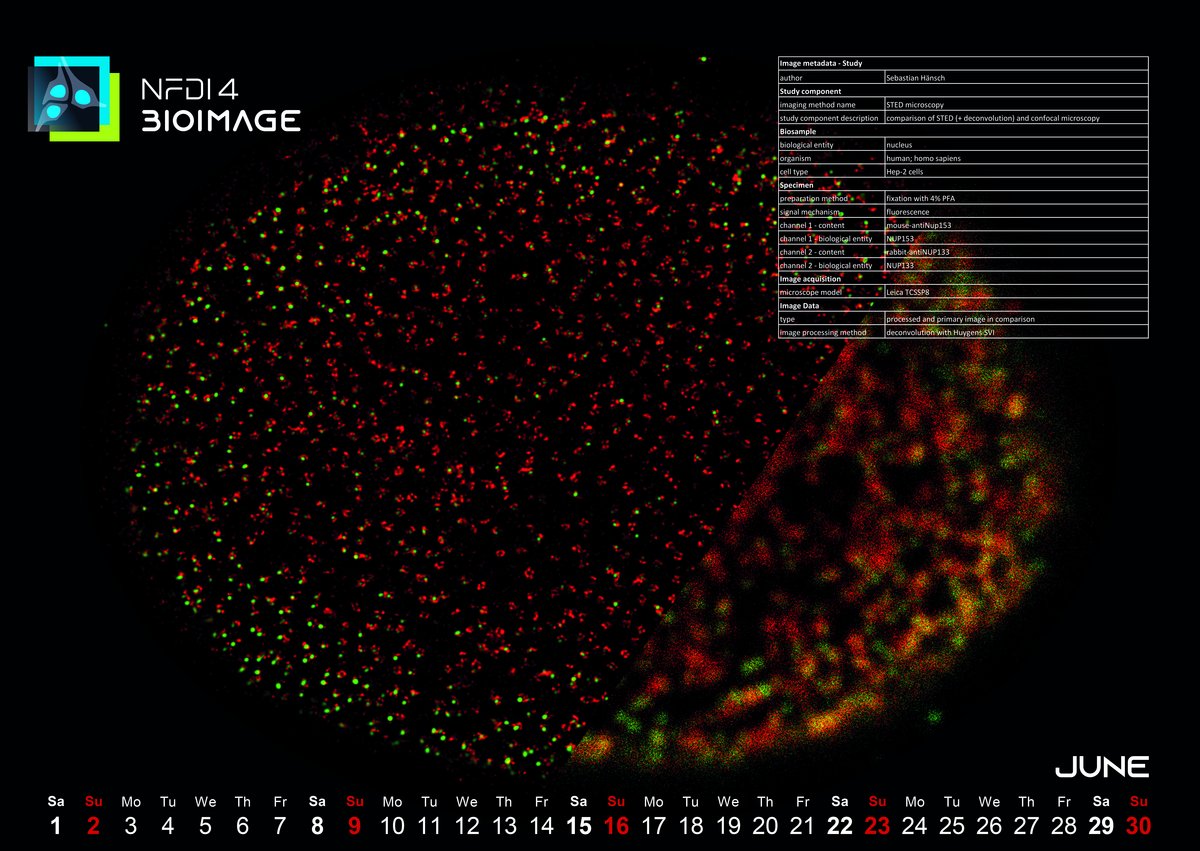
June 2024: Resolution comparison of confocal microscopy versus STED (+deconvolution)
The image for this month was captured in 2016 during Dr. Sebastian Hänsch's doctoral research at the Center for Advanced Imaging (CAi) at Heinrich-Heine-University in Düsseldorf. Sebastian mainly deals with fluorescence microscopy spectroscopy and super-resolution microscopy. The image depicts stained nuclear pore complexes, which Sebastian utilized as a standard control for benchmarking the performance of an STED system. Notably, the red fluorescence highlights the ring-like structure of the nuclear pore complex protein Nup133 (NUP133), discernible only at a certain level of resolution enhancement. This served as an initial quality check to ensure the system was functioning optimally at the onset of the measurement session. Deconvolution techniques were employed to enhance image quality, a common practice in imaging datasets. Additionally, comparing the image with confocal microscopy provided valuable insights into the extent of resolution improvement achieved by STED.
The question is whether the image is also Findable, Accessible, Interoperable, and Reusable (FAIR)? Sebastian says: “The image is recorded back days when there was not that much of FAIR principles or proper documentation. Now for initial estimations like that, we at least use templates or stick to a few default parameters for the image acquisition. But working on FAIR for images like control images makes sense, so we are working on that in at CAi.”
The increasing complexity and volume of biological imaging data, reflecting trends seen in other fields, pose significant challenges for Research Data Management (RDM). This highlights the importance of ensuring that image data adhere to the FAIR principles. The significance of RDM in microscopy and bioimage analysis has been underscored by NFDI4BIOIMAGE, a national initiative funded by the Deutsche Forschungsgemeinschaft (DFG). Operating within Germany's National Research Data Infrastructure (NFDI), this initiative collaborates to address the critical issue of data management for bioimages, emphasizing the necessity of FAIR principles in microscopy and bioimage analysis.
To ensure successful engagement and standardization within the user community, adherence to the REMBI guideline, or Recommended Metadata for Biological Images, is crucial. This guideline outlines the minimum information necessary for biological images. Additionally, utilizing an open-source platform like Open Microscopy Environment Remote Objects (OMERO) serves as a valuable Research Data Management (RDM) tool for both short-term and long-term use of imaging data. OMERO facilitates streamlined access to stored image data, enhancing collaboration and ensuring efficient management of biological images.
Keep following us for more coherent implementation of the FAIR principles in bioimaging which is a key goal of our consortia NFDI4BIOIMAGE.
Dr. Mohsen Ahmadi, Data Steward in the NFDI4BIOIMAGE consortium and Leibniz Institute for Plasma Science and Technology - INP Greifswald
