March 2024: Dinner for Whom?
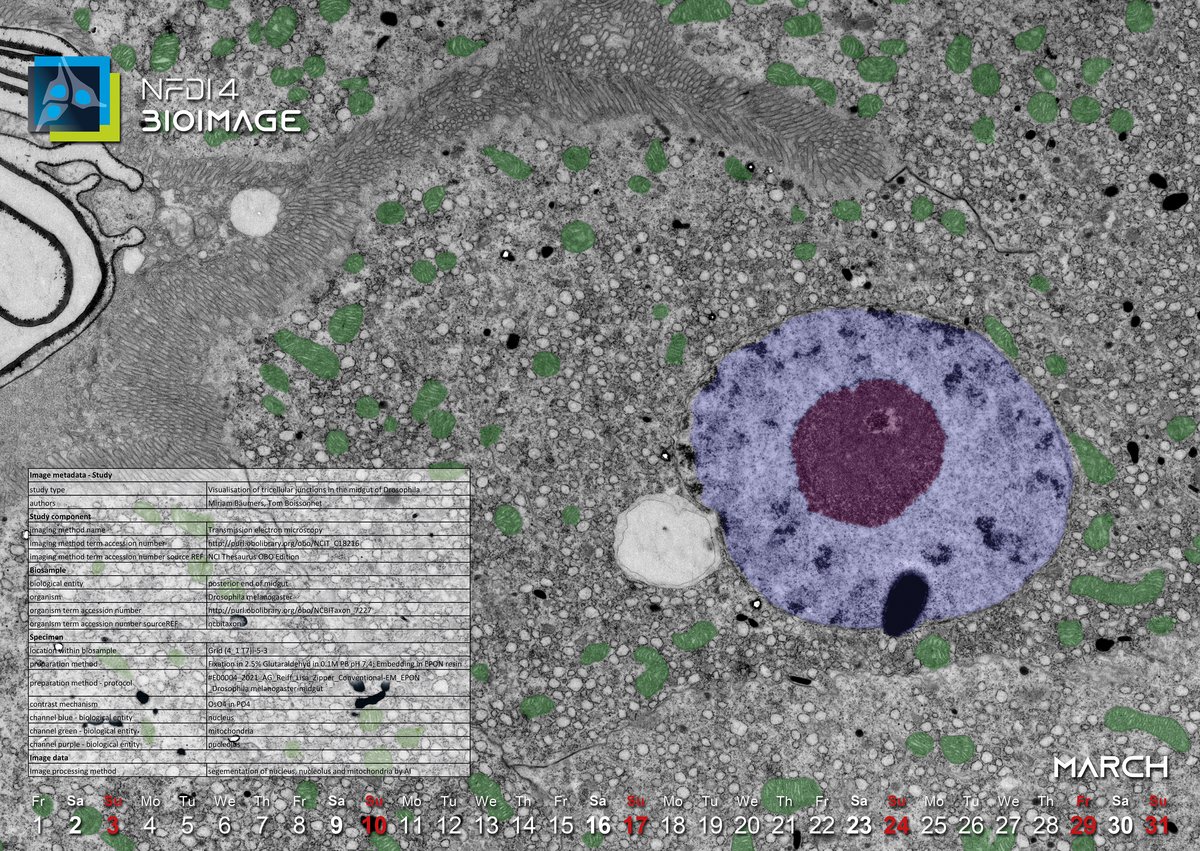
This month’s image depicts a section of the posterior end of a Drosophila melanogaster’s midgut in a transmission electron microscopy micrograph, with an artificially colored overlay of the nucleus (blue), nucleolus (purple), and mitochondria (green) resulting from an artificial-intelligence assisted segmentation. The image was imported into OMERO, a software platform for visualizing, managing, and annotating metadata of scientific images, and loaded straight into QuPath via an OMERO plugin, where the SegmentAnythingModel (SAM) was set up for segmentation. From box inputs, SAM detected the regions of interest (ROI) that we uploaded back to OMERO as annotation of the original image, waiting for further review, analysis or montage. Via OMERO, the image can be made FAIR (What is FAIR? See January and February 2024).
But what goes beyond FAIR? When acquiring any type of data, it is desirable to comply with the FAIR principles to make data reusable. However, it is equally important to make the (meta)data "fit for purpose." In other words, while following these principles to make your (meta)data FAIR, you should also think about who should be able to reuse the data. Because from the user's perspective, the quality of (meta)data is context-dependent. It can be of high quality in one context and of (relatively) poor quality in another. Let's go back to our image from this month: what is the scientific context of our image, and what would be the purpose of reusing it? Technically improve a microscopic method? - In this case, the focus of the metadata description would be on the technical part of the image capture. Or is it to find out what our Drosophila melanogaster ate for dinner? - In this case, the metadata description would focus on the examined object and the experimental conditions.
In our consortium, we are working on solutions and workflows to make this seemingly tedious metadata annotation as simple and automated as possible and to make your data FAIR and fit for multiple purposes. This includes the automated import of multimodal and harmonized metadata, the conversion of proprietary data formats into a universal data format, and the provision of the technical infrastructure.
Keep following us for more!
Maximilian E. Müller, Data Steward in the NFDI4BIOIMAGE consortium (Team Open Science, KIM, University of Konstanz)
